Lab mechanics (how to work through the labs of this course)¶
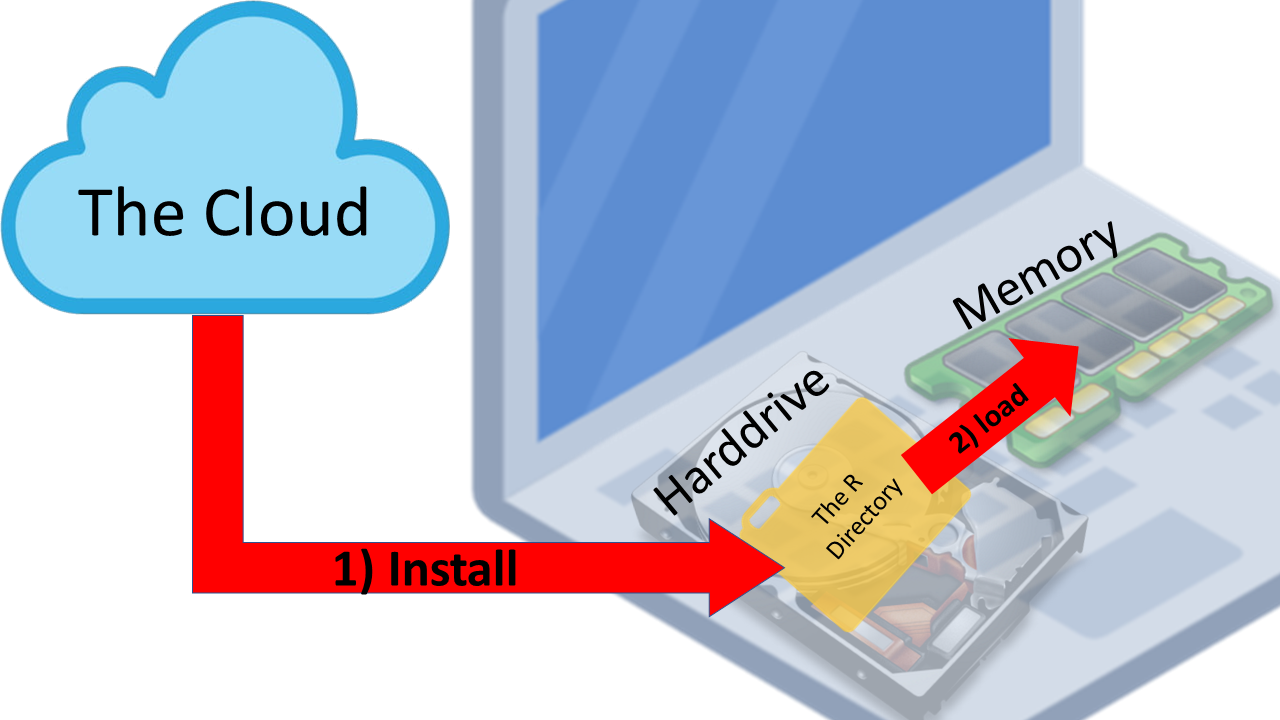
For the labs in this course you will need 3 programs:
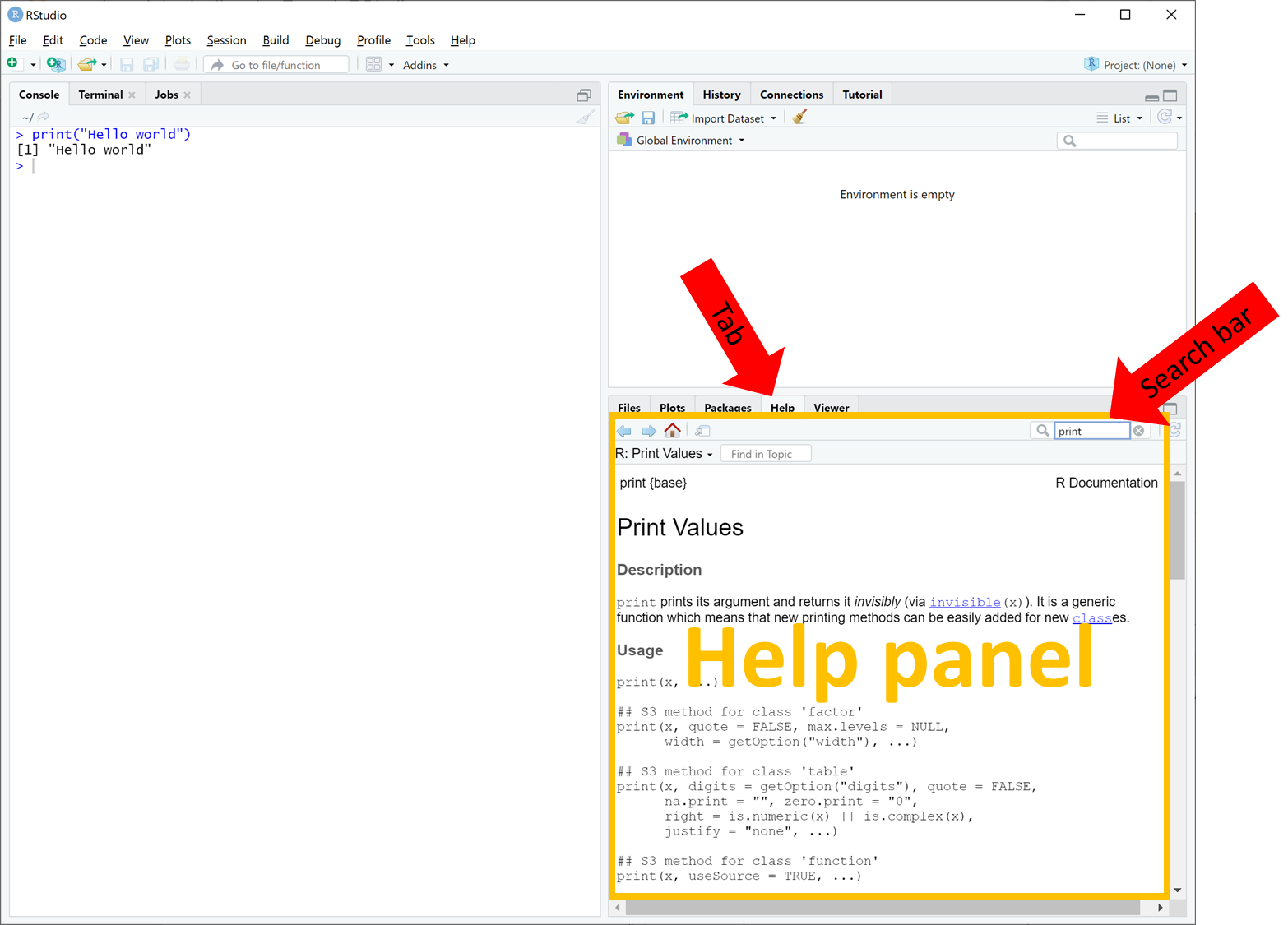
- Lab manual: It is a web-page with instructions on what to do in the lab. If you are reading "this", you already found the lab manual. The Lab manual is viewed inside your browser (e.g. Firefox, Chrome, Safari, etc.).
- Brightspace LAB Questions: In the Lab manual there are several questions that you will need to answer in a corresponding Brightspace LAB Questions. Same as the Lab manual, the Brightspace LAB Questions is viewed inside your browser (e.g. Firefox, Chrome, Safari, etc.). You can get to the Quizzes section in Brightspace following Assessment > Quizzes
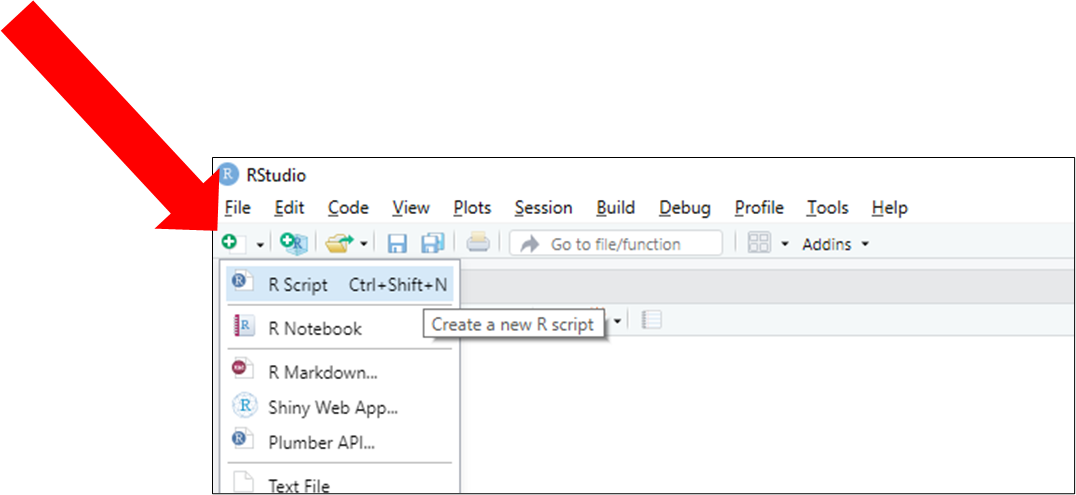
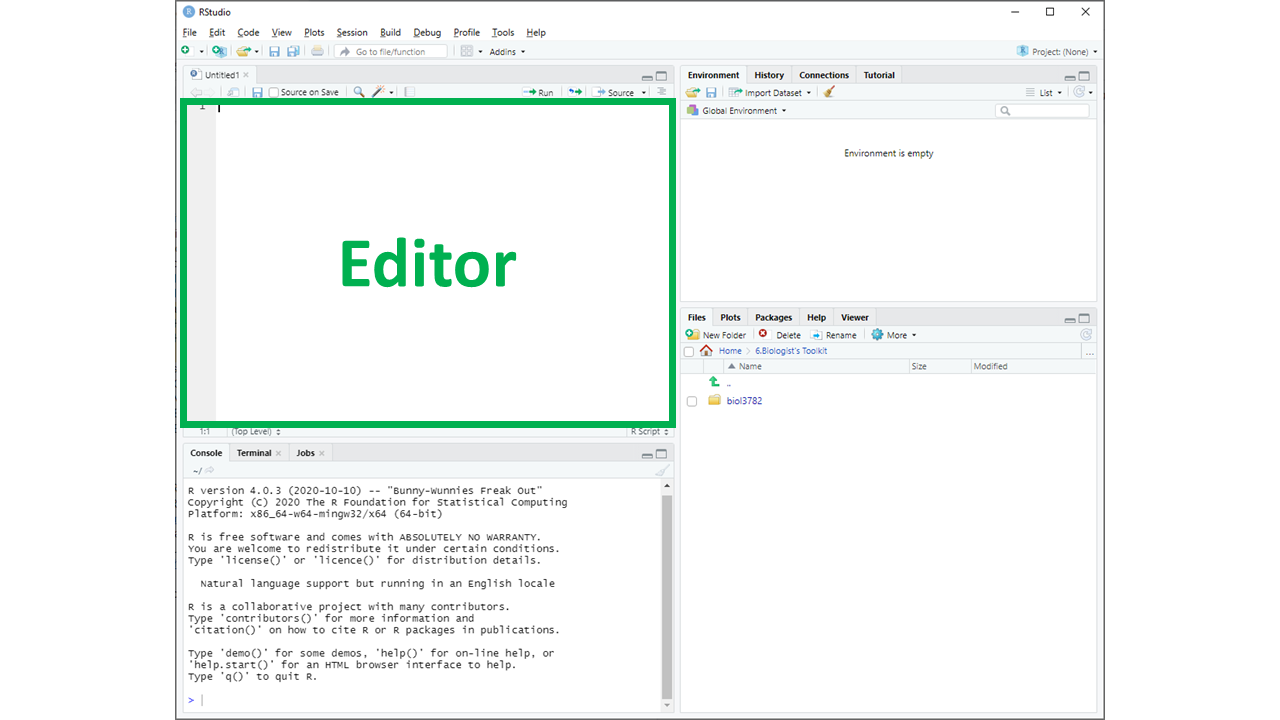
- RStudio: This is the main program where you will use to write and run R code. We'll talk more about RStudio below.
Work flow in a typical Lab¶
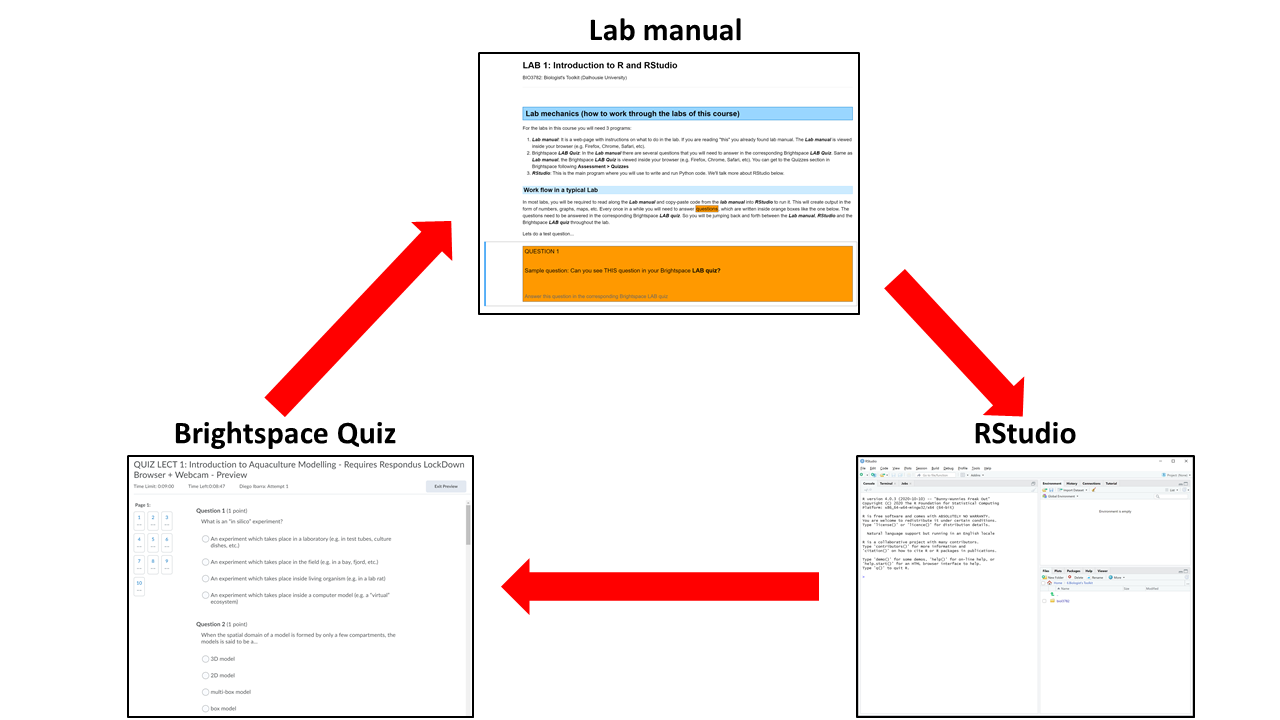
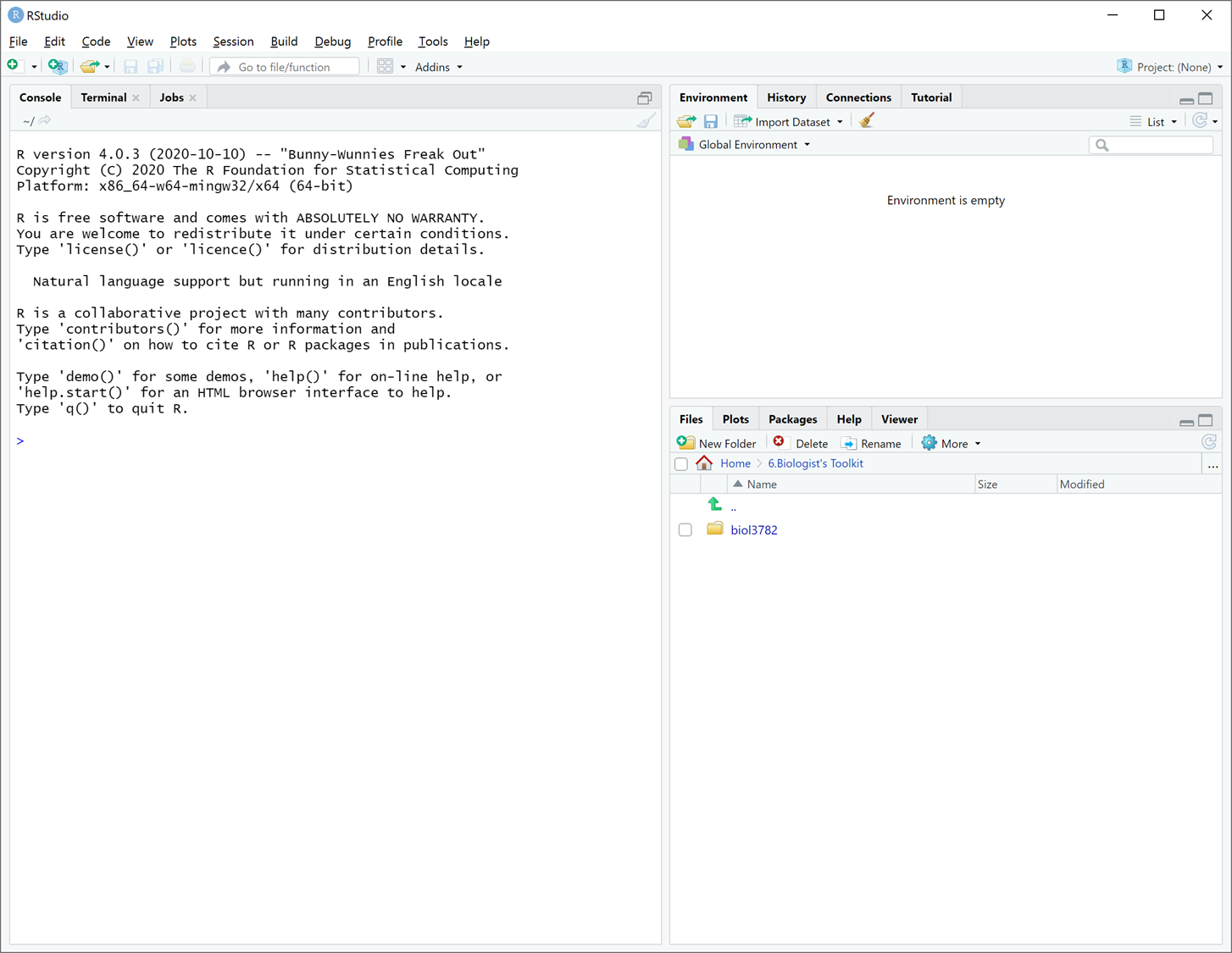
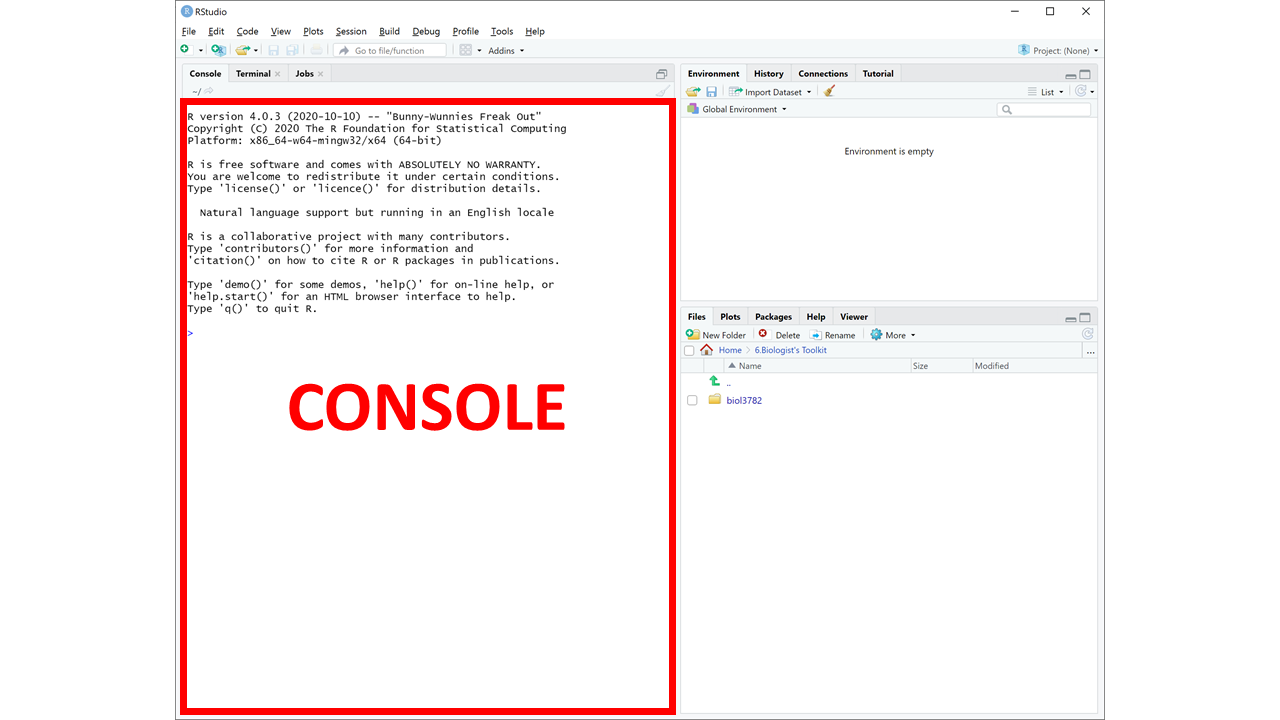
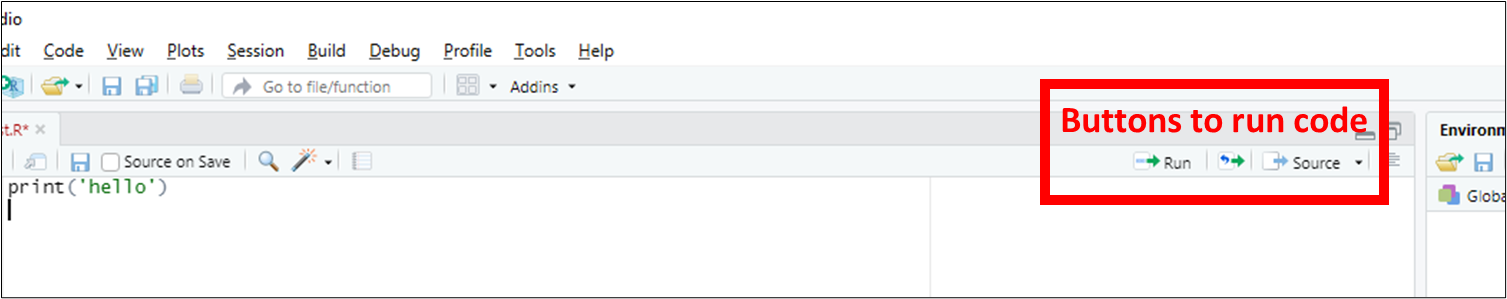
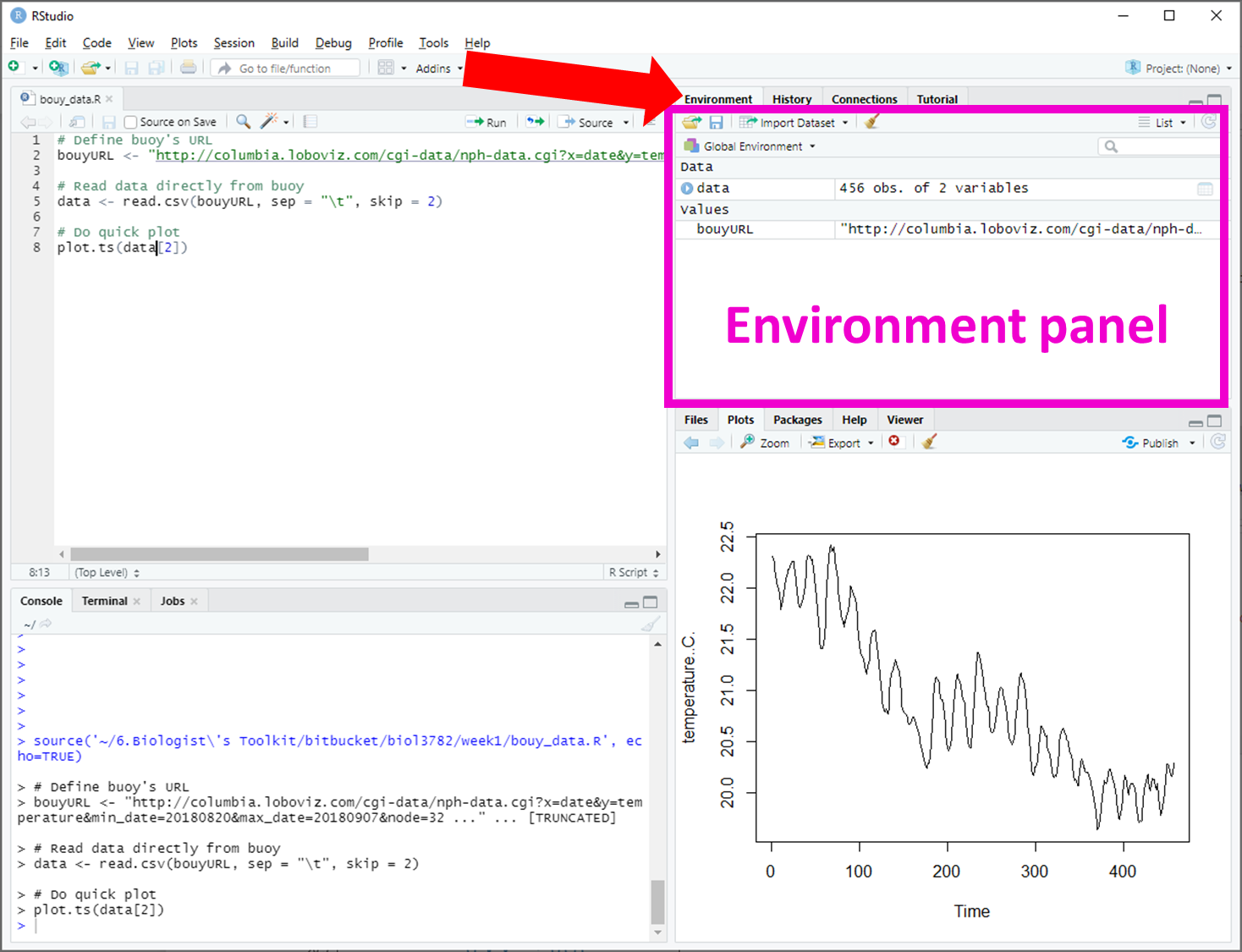
In most labs, you will be required to read along the Lab manual and copy-paste code from the lab manual into RStudio to run it. This will create output in the form of numbers, graphs, maps, etc. Occasionally, you will need to answer questions, which are written inside orange boxes like the one below. The questions need to be answered in the corresponding Brightspace LAB Questions. So, you will be jumping back and forth between the Lab manual, RStudio and the Brightspace LAB Questions throughout the lab.
Let's do a test question...


 LOBO Buoy
LOBO Buoy Columbia River (Oregon, USA)
Columbia River (Oregon, USA)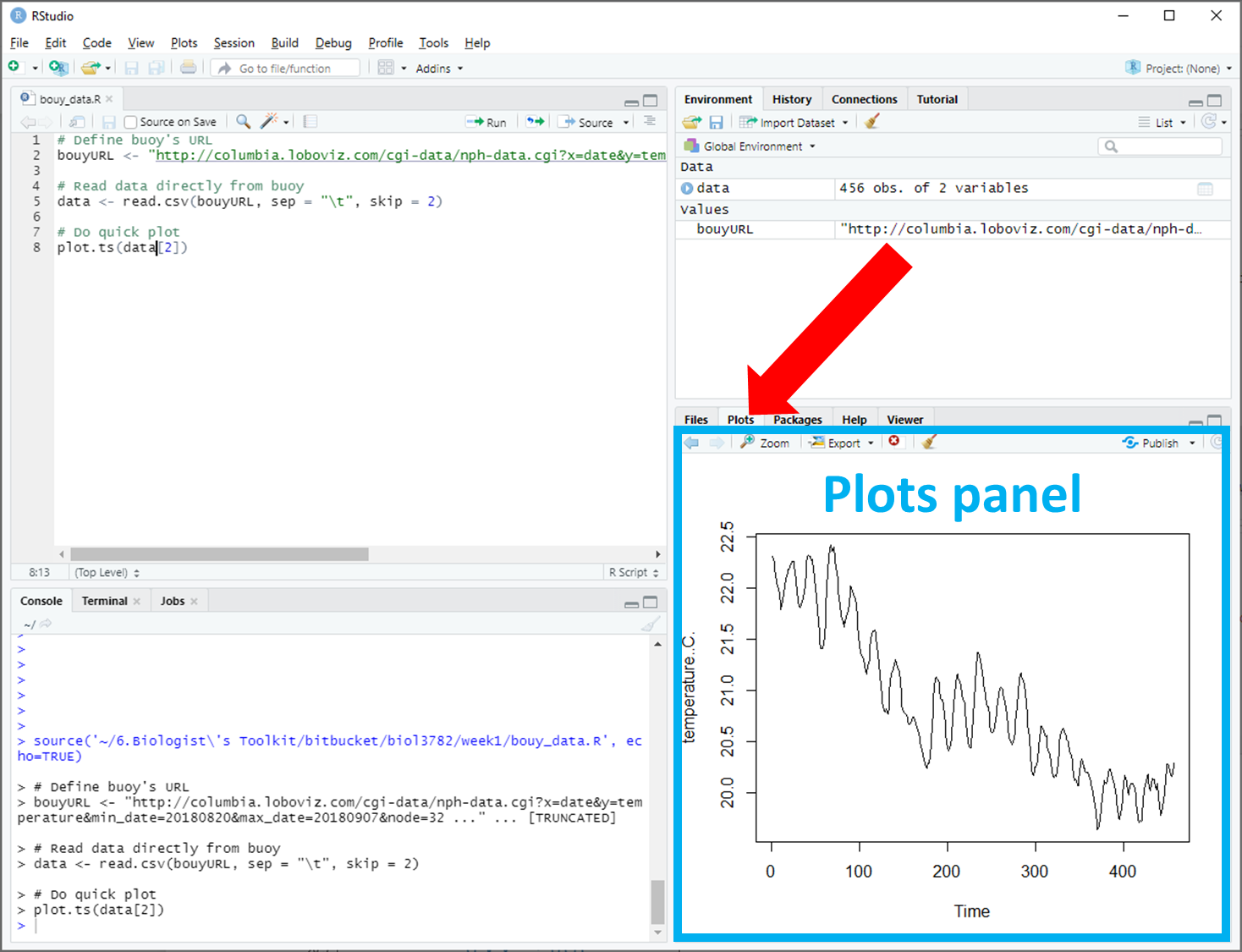
 Slocum Glider
Slocum Glider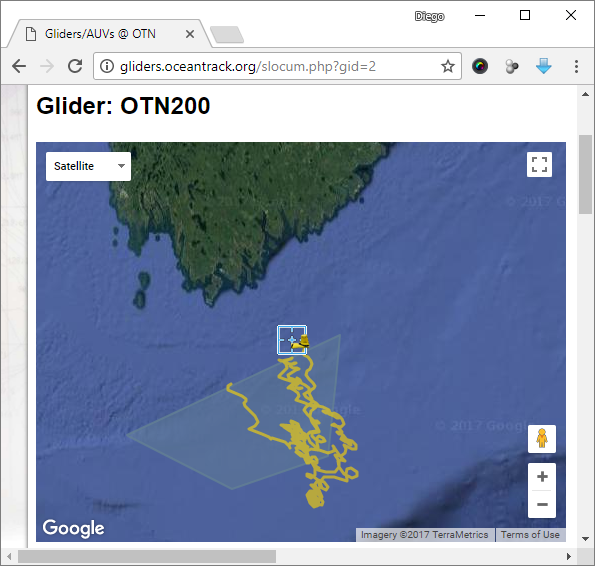 Current deployment
Current deployment